Researchers from the University of Queensland have created a detailed atlas showing how human pluripotent stem cells develop into many different cell types. Using single-cell RNA sequencing, the team followed more than 60,000 individual cells over time as they transitioned from a pluripotent state (where cells can become any type of cell in the body) to committed, specialized cells representing all three germ layers: ectoderm, mesoderm, and endoderm.
Experimental design and rationale
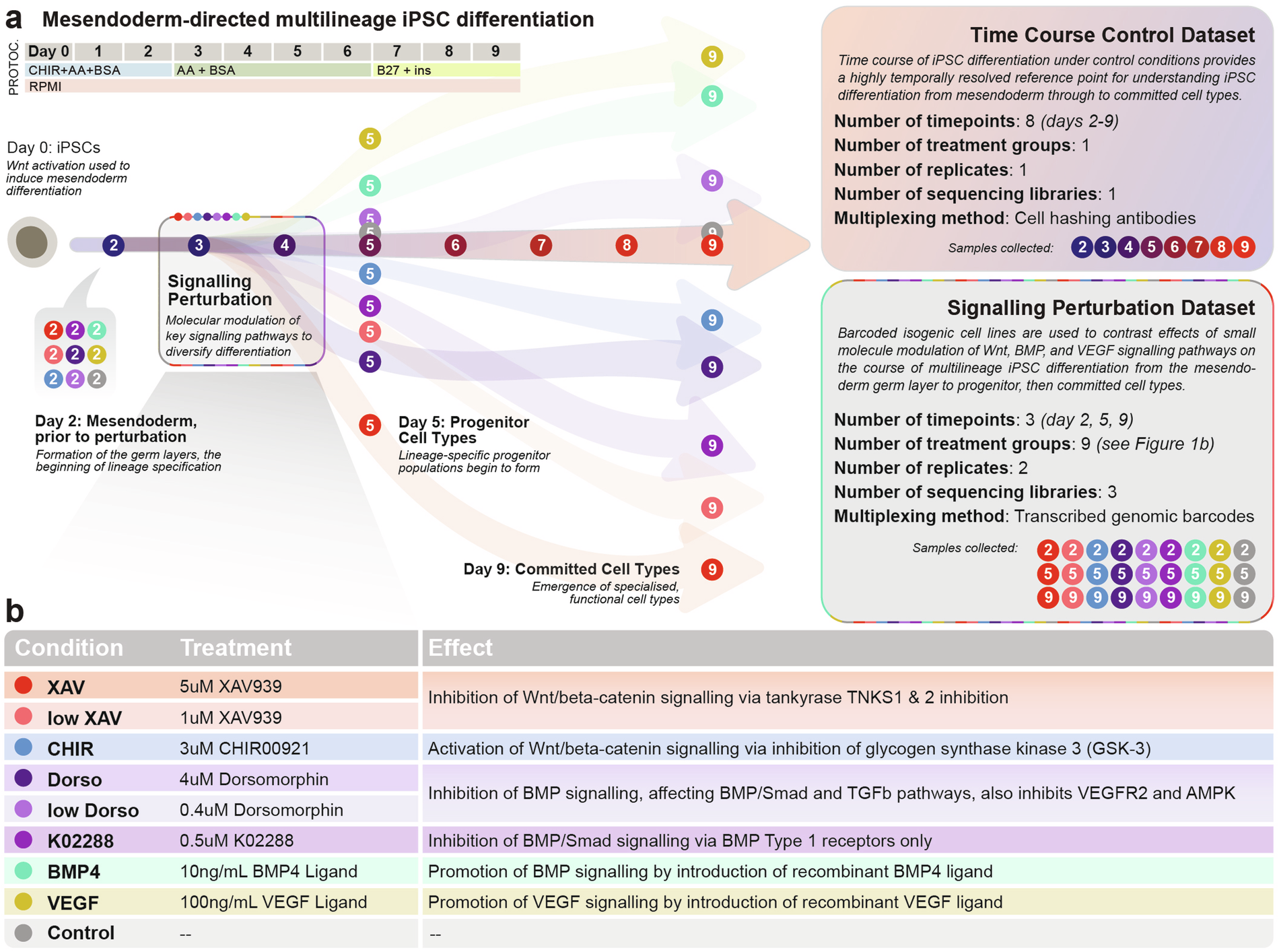
(a) High resolution time course dataset and small molecule signalling perturbation datasets were designed to complement each other as reference points for multilineage differentiation from pluripotency in vitro. Base differentiation protocol used in the time course dataset and added to for the signalling perturbation dataset is shown on the top left. Protoc.: Protocol; CHIR: CHIR00921; AA: Ascorbic Acid; ins: insulin. See Methods for more protocol details and Table 1 for details about time point/replicate/sequencing library assignment for each sample in the sequencing perturbation dataset. (b) Description of the molecules used in the signalling perturbation dataset.
This time-course dataset provides a new way to study how key developmental signals influence cell fate decisions. Until now, most high-resolution transcriptomic studies have focused on in vivo development, but there has been a lack of similar data for cells differentiating in lab-grown conditions. This work fills that gap and offers a valuable reference for comparing in vitro stem cell models with natural embryonic development.
These insights have major implications for regenerative medicine, drug screening, and the development of better cell differentiation protocols. By understanding how gene expression shifts during the transition from stem cell to specialized cell, scientists can improve the precision of protocols used to generate specific cell types for therapy or research.