DNA-based genetic markers have been used in a wide range of biological fields, including medicine, genetics, and molecular biology. Genotyping is essential for these research, and various methods have been devised [13, 25].
The first generation of genotyping was using restriction fragment length polymorphism (RFLP) or random amplified polymorphic DNA (RAPD) markers [1]. These markers are not frequently in the genome, and the experiments were laborious and time-consuming. In the next stage, genotyping by PCR and electrophoresis using simple sequence repeat (SSR) markers has been commonly performed since 1990’s [18, 26, 31]. Although it has led to many genetic achievements, it is now considered labor intensive in the age of next-generation sequencing (NGS). High-throughput NGS genotyping methods are now available. Because whole-genome sequencing for all genetic analysis populations is expensive, it is common to produce a reduced representation of the genome before sequencing [13]. There are several methods to produce a reduced representation of the genome, such as RAD-seq [3], GBS [16], MIG-seq [32], and GRAS-Di® [17]. However, these methods do not allow prior knowledge of marker locations. SNP genotyping assay using microarray technology can solve this problem [21]. It is commercially available [2], but the cost of design novel SNP array is high compared to the NGS-based methods mentioned above. Targeted amplicon sequencing (TAS) is also a promising method. For genotyping by TAS, the sequences around the targeted polymorphisms are amplified by multiplex PCR, and the amplicons are sequenced via next-generation sequencing (NGS). Therefore, markers can be designed at desired positions. The concept of TAS was first reported more than a decade ago [6]. Similar methods have been reported under different names, such as Genotyping-in-Thousands by sequencing (GT-seq) [9] and Multiplex PCR Targeted Amplicon Sequencing (MTA-Seq) [29]. It is also provided commercially as Ion AmpliSeq technology (Thermo Fisher Scientific, Inc.) [30].
In crop science, bi-parental quantitative trait loci (QTL) analyses are often conducted to identify genomic regions related to agronomically important traits. For this application, it is important to ensure enough markers across the genome while keeping costs as low as possible. TAS has a potential to meet this requirement. However, TAS is currently not frequently used for this application for several reasons. First, a reference genome is required for custom primer design. Second, it is expensive to purchase hundreds or thousands of primers. Third, it requires advanced knowledge and skills in bioinformatics. In recent years, the first problem is not so critical because reference genomes have been established for many crop species [34]. Natsume et al. [28] showed that the second and third problems could be overcome by using cheaper low-concentration mixed primers and developing a free software named V-primer.
The workflow of TAS is roughly divided into three steps (Fig. 1). First, DNA markers are extracted from the NGS data of the parental lines, and primers are designed. Second, the NGS library is constructed using the designed primers, and sequencing is performed. Third, the data obtained by NGS are analyzed, and the genotypes at each DNA marker of each line are determined. The first and third steps require specialized bioinformatics analysis.
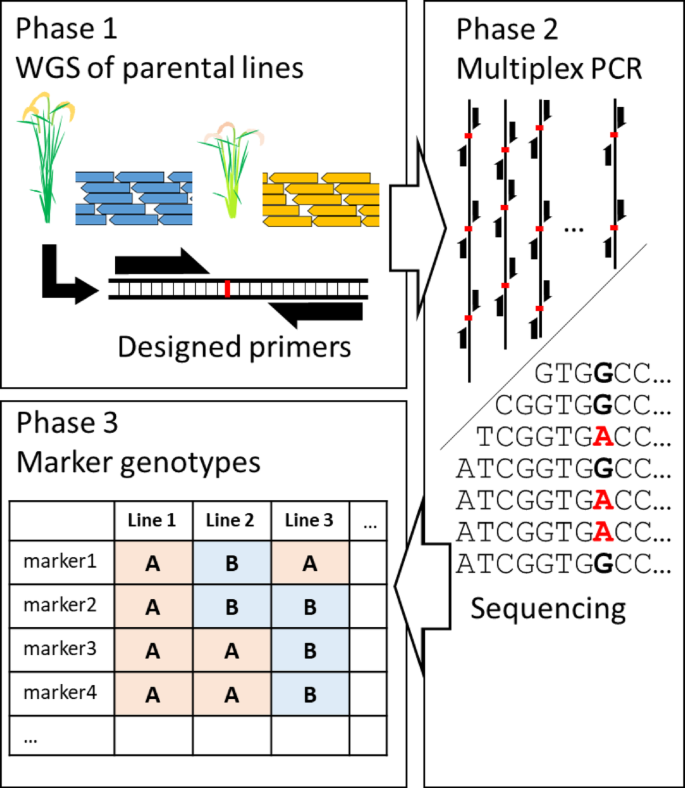
Schematic diagram of genotyping by targeted amplicon sequencing workflow
V-primer [28] can perform the first step, DNA marker extraction and high-throughput primer design, at once. However, no integrated pipelines have been developed to handle the third step, analysis after sequencing. There is also room for improvement in primer design strategies. To make TAS more generally available, it is necessary to establish user-friendly pipelines that covers whole the experiments.
In this study, we developed a novel genome-wide primer design tool, MKDESIGNER, which implements specialized functions for genotyping by TAS. We also developed the post-sequencing analysis tool TASEQ. This tool is a pipeline that takes FASTQ files as input and outputs genotype files in a format that can be used directly for QTL analysis. By implementing these tools, genotyping by TAS for populations which are derived from cross among two to several fixed parental lines can be done easily.
In this paper, we explain how MKDESIGNER and TASEQ work and provide a practical example of genotyping by TAS in rice using these tools.
Implementation
Overview of the tool
MKDESIGNER and TASEQ are command line interface tools that have been verified to work on Ubuntu 20.04 and later. The source codes are written in Python and are available at GitHub (https://github.com/KChigira/mkdesigner, https://github.com/KChigira/taseq)
They can be installed via Bioconda, including their dependencies. The workflow is shown in Fig. 2. MKDESIGNER has three commands, and TASEQ has four commands. The role of each command and the external tools used are described in the following sections.
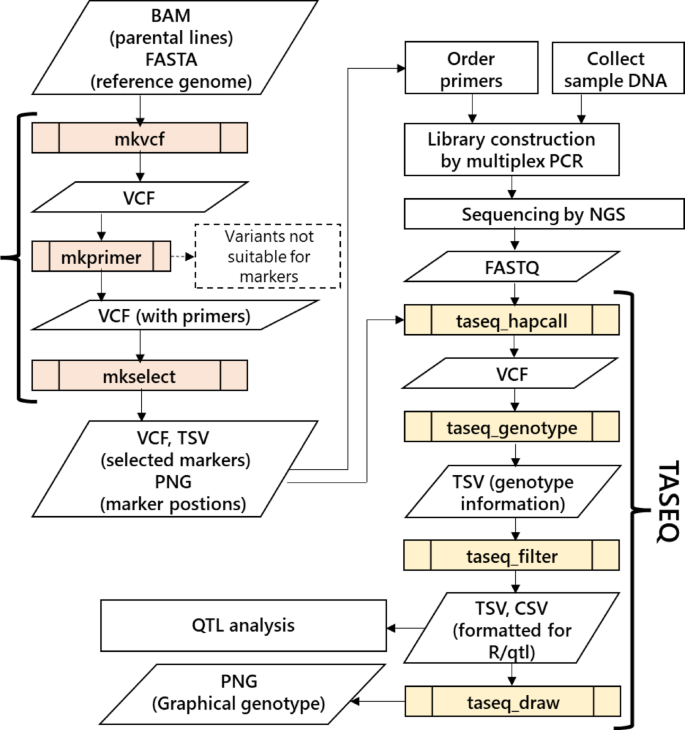
Workflow of genotyping by TAS with automated data analysis using MKDESIGNER and TASEQ
‘mkvcf’
This command is responsible for creating a VCF file from the NGS data of the parent varieties. It requires BAM-formatted NGS data from two or more parental lines and a FASTA-formatted reference genome as input. This command produces a VCF file using GATK HaplotypeCaller [27] and BCFtools [12]. Pre-created VCF files can also be used for subsequent analysis, but they may lack the information necessary for primer design (ex. sequence depth of each polymorphism). Therefore, we provide a command to create a VCF in a format suitable for subsequent analysis.
‘mkprimer’
This command is responsible for designing primers that amplify around polymorphisms. To prepare markers, polymorphisms suitable for primer design are selected from the input VCF table according to the following criteria:
-
(1)
Genotypes differ between parental lines. For example, the GT fields of parental lines A and B in the input VCF are ‘0/0’ and ‘1/1’, respectively.
-
(2)
The reliability of polymorphism calling is high (passes GATK VariantFiltration: QD < 20.0 and FS > 200.0 and SOR > 10.0, fixed values).
-
(3)
The sequence depth is within the specified range (default: 2 ~ 200, modifiable).
The primers used were designed using Primer3 software [33]. The primers used are designed so that they do not overlap other DNA mutations and are in accordance with other specified conditions (Supplementary Fig. 1). The designed primers were checked to determine whether their amplicons were specific to the genome via BLAST software [8]. The BLAST condition is based on the settings described in the report of Primer-BLAST software [35]. By default, ‘mkprimer’ explores as many markers as possible (Fig. 3a). If users want to save time, the number of polymorphisms to search for primers can be reduced by selecting the appropriate options. Finally, a VCF-formatted file containing the sequences of the designed primers added to the ‘INFO’ column is output.
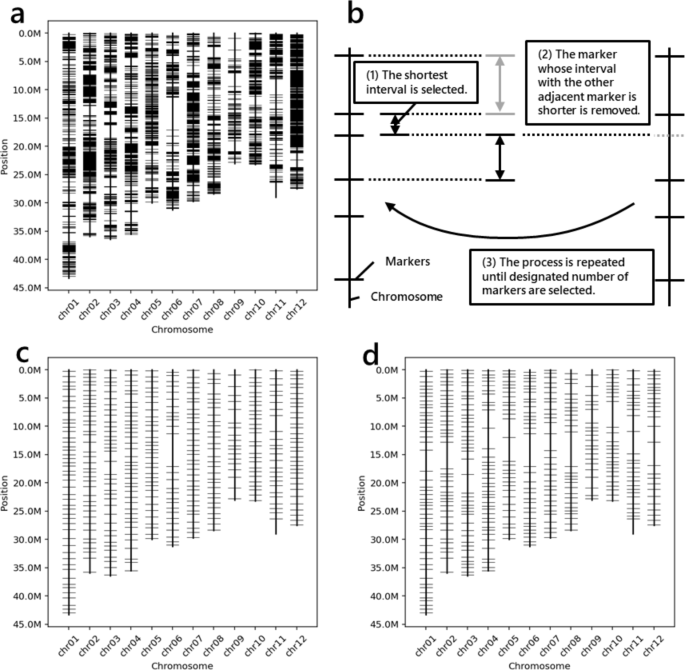
Example for the output of MKDESIGNER. a Genome-wide markers made by the ‘mkprimer’ command. b The strategy selecting markers in the ‘mkselect’ command. a Normally selected 384 markers using the ‘mkselect’ command. d 384 markers selected by the ‘mkselect’ command using the ‘–density’ option to reduce markers near the centromeres
‘mkselect’
This command narrows the markers to a specified number at equal intervals. The ‘mkselect’ works according to the following algorithm (Fig. 3b):
-
(1)
The physical distance between all the DNA markers is calculated, and the pair of markers with the narrowest spacing is identified.
-
(2)
For that pair of markers, the physical distance to the other adjacent markers is calculated. The marker with the smaller distance is removed.
-
(3)
These steps are repeated until the specified number of markers is reached.
The output files are a VCF file containing only the selected markers, a TSV file containing primer information, and a PNG file illustrating the physical positions of the selected markers (Fig. 3c).
In linkage mapping, physical distances among markers are not proportional to genetic distances, especially near centromeres. ‘mkselect’ can adjust the marker density of such regions by adding a tab-delimited file of the specified format to the ‘–density’ option (Fig. 3d).
`taseq_hapcall’
The following commands belong to TASEQ, which is responsible for post-sequencing analysis. The graphical workflow is provided in Supplementary Fig. 2. The first command is responsible for extracting target polymorphisms from raw sequence data (FASTQ) of multiplex PCR amplicons. First, the sequence reads in the input FASTQ files are trimmed using Trimmomatic [4] (a). Moreover, a list of sequences before and after the target polymorphism was generated using SAMtools in FASTA format (b). Second, the reads in (a) are mapped to (b) using BWA [24]. Third, the genotypes of the markers are determined using GATK HaplotypeCaller. Finally, the results for all the lines are combined and output as a VCF file.
‘taseq_genotype’
This command determines which genotype of each line, and each marker is homozygous for parent A (A), homozygous for parent B (B), heterozygous (H), or missing (-). The algorithm proceeds as follows:
-
(1)
Markers with a sequence depth less than 10 (default, modifiable) are considered missing (-).
-
(2)
If the proportion of minor alleles is less than 10% (default, modifiable), the marker genotype is homozygous for the major allele (A or B).
-
(3)
If the marker genotype is not determined in (2), the chi-square value is calculated for the ratio of the number of reference alleles and alternative alleles. If the p-value of chi-squared test is greater than 0.05 (modifiable), the marker genotype is missing (-); otherwise, it is heterozygous (H).
The genotypes of each line and marker are output as a TSV file.
‘taseq_filter’
This command removes markers with a specified percentage of missing data or less than a specified minor allele frequency. If parental lines are included in genotyping, only markers that are supported by the marker genotypes of the parental lines can be selected. The output CSV file is formatted to be used directly as genotype data in R/qtl [5].
‘taseq_draw’
This command visualizes marker genotypes at the chromosomal level for each line. It outputs PNG files for the number of lines in the output directory.