Researchers at The Hospital for Sick Children in Toronto have demonstrated the significant benefits of using RNA sequencing (RNA-seq) for cancer diagnosis and treatment decisions. While most cancer diagnostics traditionally focus on DNA sequencing to identify mutations, RNA sequencing offers unique advantages by analyzing gene expression and detecting fusion genes and splice variants that DNA tests may overlook.
In their comprehensive clinical study, the team applied a single targeted RNA-seq assay to over 2,300 tumor samples from children and adults, covering a wide range of cancers including solid tumors, central nervous system cancers, and blood cancers. Many of these samples were formalin-fixed and paraffin-embedded, which often presents challenges for molecular testing, yet the RNA-seq assay demonstrated a low failure rate of just 4.8%. Impressively, the test provided meaningful molecular information for 87% of patients.
Clinical impact of targeted RNA-seq
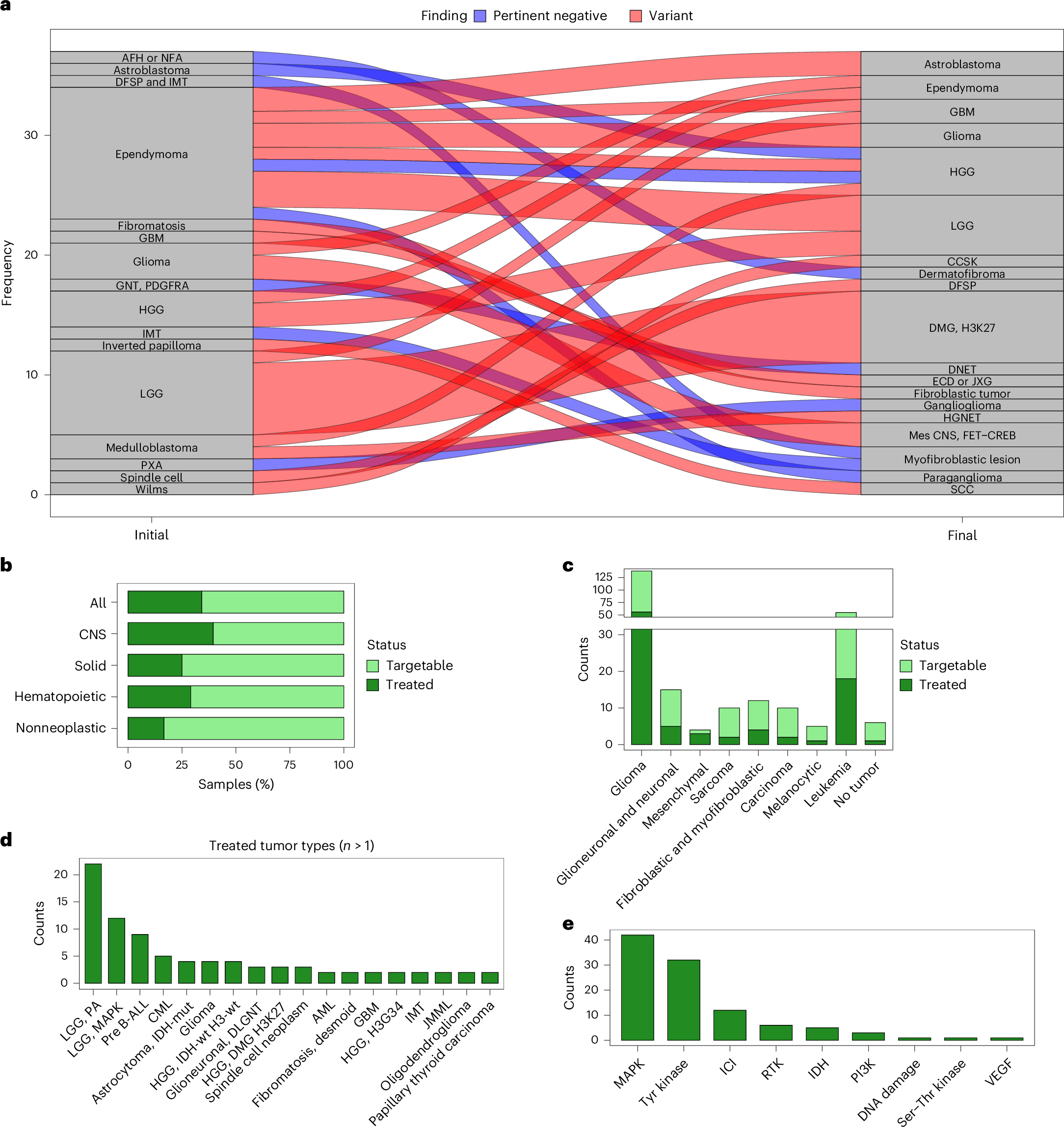
a, Alluvial plot showing the numbers of tumors re-diagnosed following targeted RNA-seq and colored according to whether RNA-seq identified diagnostic alterations (red) or pertinent negative findings (blue) to rule in or out specific diagnoses. AFH, angiomatoid fibrous histiocytoma; CCSK, clear cell sarcoma of the kidney; DFSP, dermatofibrosarcoma protuberans; DMG, H3 K27, diffuse midline glioma, H3 K27-altered; ECD, Erdheim–Chester disease; GBM, glioblastoma, IDH-wt; GNT, PDGFRA, myxoid glioneuronal tumor, PDGFRAp.K385‐mutant; HGG, high-grade glioma; IMT, inflammatory myofibroblastic tumor; JXG, juvenile xanthogranuloma; LGG, low-grade glioma; Mes CNS, FET–CREB, intracranial mesenchymal tumors with FET–CREB fusion; HGNET, high-grade neuroepithelial tumor; NFA, nodular fasciitis; SCC, squamous cell carcinoma; PXA, pleomorphic xanthoastrocytoma. b, Proportion of all CNS tumor, solid tumor, leukemia and nonneoplastic samples with targetable findings, where patients subsequently received targeted therapy. c, Counts of tumors of the indicated types with targetable alterations, where the patient subsequently received targeted therapy. d, Counts of indicated tumor types where patients received targeted therapy. LGG, PA, pilocytic astrocytoma; LGG, MAPK, diffuse low-grade glioma, MAPK pathway-altered; Glioneuronal, DLGNT, diffuse leptomeningeal glioneuronal tumor; HGG, DMG H3K27, diffuse midline glioma, H3 K27-altered; GBM, glioblastoma, IDH-wt; HGG, H3 G34, diffuse hemispheric glioma, H3 G34-mutant. e, Counts of indicated types of targeted therapy administered following findings from targeted RNA-seq. DNET, dysembryoplastic neuroepithelial tumor; ICI, immune checkpoint inhibitor; RTK, receptor tyrosine kinase; VEGF, vascular endothelial growth factor.
RNA sequencing revealed alterations that not only confirmed or refined initial diagnoses but also identified clinically actionable genetic changes. This led to changes in patient treatment plans, including the administration of targeted therapies tailored to the specific molecular profile of the cancer. These findings highlight RNA-seq’s value not just as a complementary method but as a powerful standalone diagnostic tool that can streamline testing workflows.
Beyond its diagnostic power, RNA sequencing also offers practical advantages. It requires less tissue material, which is important when biopsy samples are limited, reduces the time to get results, and helps control costs. For clinicians and patients alike, this means quicker, more accurate diagnoses and more personalized treatment options that could improve outcomes.
This work from The Hospital for Sick Children supports a growing movement toward incorporating RNA sequencing into routine clinical practice for cancer diagnostics. By capturing a broader spectrum of molecular changes than DNA-based methods alone, RNA sequencing helps clinicians better understand tumor biology and make more informed treatment decisions.