Diabetic retinopathy detection framework
As depicted in Fig. 3, the Guided Progressive Multi-scale KL-Ensemble Network (GPMKLE-Net) is introduced as an innovative framework specifically tailored for the multi-class classification of diabetic retinopathy severity. GPMKLE-Net seamlessly integrates randomized multi-scale image reconstruction, ensemble learning with KL-divergence-based collaborative regularization, and a guided learning approach, leveraging the meta-learning concept of self-paced progressive learning to optimize performance in few-shot medical image classification tasks. This method enables the model to systematically evolve from recognizing easily identifiable pathological features to mastering more complex and subtle ones, significantly enhancing its efficiency in handling limited training datasets. Our findings highlight the potential of GPMKLE-Net to revolutionize DR severity classification, paving the way for more accurate and efficient medical image analysis.
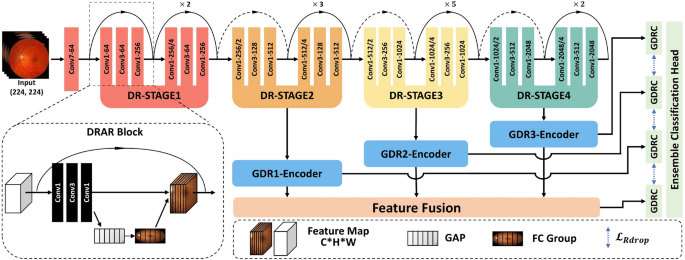
This figure comprehensively illustrates the architectural design of GPMKLE-Net, a network specifically designed for detecting diabetic retinopathy (DR). It showcases the overall structure, featuring several key components: the Diabetic Retinopathy Attention Residual (DRAR) Block for feature extraction, various stages of the Diabetic Retinopathy Network Backbone (DR-STAGE), the Guided Diabetic Retinopathy Encoder (GDR-Encoder), the multi-scale feature fusion layer, and the ensemble learning classification module with Kullback-Leibler (KL) divergence regularization. The input image passes through the network backbone, where it captures and emphasizes lesion features at multiple scales across different stages. These features are then re-encoded by the GDR-Encoder. Subsequently, through feature fusion and an ensemble learning classification algorithm enhanced with KL divergence regularization, features extracted in previous stages are effectively integrated to determine the DR grade of the input image.
Backbone network
The backbone of GPMKLE-Net employs ResNet-50, a widely used convolutional neural network for feature extraction. In order to effectively leverage multi-stage features, the original ResNet-50 architecture was modified such that feature maps from its final three stages are extracted and subsequently integrated into the feature extraction layer of the Guided Diabetic Retinopathy (GDR) Encoder. This approach maintains the computational flow while enhancing the model’s ability to capture features at different scales.
DRAR block: DR attention residual module
Prior research has demonstrated that shallow neural networks excel at capturing texture, shape, and other fundamental features, thereby preserving essential spatial information. However, as network depth increases, there is a tendency for certain spatial details to be lost, resulting in the extraction of more abstract representations26. To address this limitation and retain the benefits of shallow feature extraction, the backbone block of our neural network has been augmented through the integration of the Squeeze-and-Excitation (SE) attention mechanism27. Within the SE module, global average pooling is first employed to aggregate spatial information across each feature map channel, effectively capturing global contextual information. This is followed by a fully connected–ReLU–fully connected structure, through which the channel-wise dependencies are learned and non-linear interactions are modeled. Ultimately, the resulting channel weights are projected back onto the original feature maps, allowing the network to selectively emphasize or suppress specific channels. By adaptively recalibrating channel-wise feature responses, the SE module enables the network to emphasize informative pathological texture features, as depicted in Fig. 3. Furthermore, the application of multi-scale convolution allows the model to attend more precisely to pathological regions, a strategy that is consistent with the findings reported by Gangwar et al. in the context of diabetic retinopathy analysis15. Through these architectural enhancements, it is anticipated that the network will be better equipped to capture subtle yet clinically significant features within retinal images.
GDRC: guided diabetic retinopathy classifier
A central function of the GDRC structure lies in its ability to integrate information derived from multiple stages of both the backbone network and the DR-Stage. Notably, the features to be fused within the GDRC are extracted by the Guided Diabetic Retinopathy (GDR) Encoder. Within the GDR Encoder, features from each stage are first subjected to a series of operations, wherein two consecutive convolutional block sequences (Convolution-BatchNorm-ReLU sequences, CBRs), each followed by max pooling, are employed. By means of these CBRs, not only is feature extraction substantially enhanced, but normalization of the intermediate representations is also facilitated, thereby ensuring that the data are optimally conditioned for subsequent fusion and processing within the GDRC.
A key aspect of the GDRC is its integration with the concept of Rdrop, which introduces diversity and robustness into the learning process. Within the GDRC, the concatenated feature maps from different stages are fed into a multi-layer perceptron (MLP). This MLP, enhanced with Dropout layers, not only adds regularization but ensures that each stage contributes unique parameters to the final prediction. By incorporating Dropout within the MLP, the GDRC effectively generates diverse sets of parameters from each stage, mimicking the behavior of multiple models. This approach not only enhances the model’s generalization capabilities but also reduces the risk of overfitting, as each “sub-model” within the GDRC contributes its own unique perspective to the final prediction. In essence, the GDRC, through its clever integration of Dropout within the MLP, transforms the concatenated feature maps from various stages into robust and diverse predictions for each DR severity category.
Ensemble classification head
In pursuit of further improvements in classification accuracy, an ensemble learning strategy is adopted. Ensemble learning has proven to be effective in enhancing model performance by combining the predictions of multiple models or, in our case, different stages of the same network.
Within our framework, each stage of the network, along with the concatenated feature stage, is regarded as an independent sub-model. In particular, the three distinct stages derived from the backbone network, together with the concatenated feature stage, collectively function as four approximate sub-models, each contributing to the overall prediction process. For every input, these four sub-models independently generate predictions regarding the severity of diabetic retinopathy. To synthesize these outputs, a voting scheme is employed, whereby the individual predictions are aggregated, resulting in a final classification that is both more robust and accurate than would be achievable by any single sub-model alone. This ensemble strategy leverages the diverse perspectives offered by each stage, thereby mitigating the risk of individual biases or errors.
Guided progressive multi-scale learning
Drawing inspiration from human cognitive mechanisms, the meta-learning paradigm derived from self-paced learning has been integrated into a progressive training methodology for deep learning systems28. This integration led to conceptualizing the Guided Progressive Multi-scale Learning (GPML) approach. The GPML process integrates two fundamental components: randomized multi-scale image reconstruction and guided loss function.
Randomized multi-scale image reconstruction
In recent years, supervised representation learning has garnered significant attention, particularly in its application to medical image analysis. Our study aligns with this trend, leveraging the advantages of multi-resolution image processing to enhance neural networks’ sensitivity to fine-grained pathological details, thereby fostering more robust and generalizable image representations. Building upon the insights provided by Du et al.28, a jigsaw generator is utilized to preprocess diabetic retinopathy (DR) fundus images, thereby facilitating the concurrent execution of supervised representation learning and multi-scale feature extraction.
In determining the optimal size and quantity of fundus image patches, careful consideration was given to both the specific requirements of the classification task and the dimensions of the training images. Throughout the training phase, multi-scale image processing was applied to each batch of fundus images, resulting in the generation of uniformly sized image patches. These patches, prior to being recombined to match the pixel dimensions of the original images, were labeled according to their respective source images. At each stage of training, patches of varying sizes were utilized to capture features at different spatial resolutions. For example, during the initial stage (denoted as (l = 1)), 64 patches were produced, each representing one-eighth of the original (224 \(\times\) 224) image, yielding patches of (28 \(\times\) 28) pixels. In the subsequent stage, 16 patches were generated, each corresponding to one-quarter of the original size ((56 \(\times\) 56) pixels). The third stage involved the creation of 4 patches, each measuring half the original dimensions ((112 \(\times\) 112) pixels). Finally, in the concatenation stage ((l = concat)), a single patch encompassing the full (224 \(\times\) 224) pixel image was employed.
The comprehensive framework for diagnosing DR, as illustrated in Fig. 4, is organized into four hierarchical training levels, corresponding to Step 1 through Step 4. Each level is designed to process and train on images of a specific scale, with each step generating its own loss feedback to guide the learning process. Features extracted from the first three steps are passed through their respective Deep-Feature Encoders and subsequently fused in Step 4 via the Multi-level Feature Fusion module. This fusion process integrates the hierarchical features from Steps 1–3, producing a unified representation in Step 4. The framework generates four feature maps: the individual outputs from the deep-feature encoders of Steps 1–3 and the fused output from Step 4.
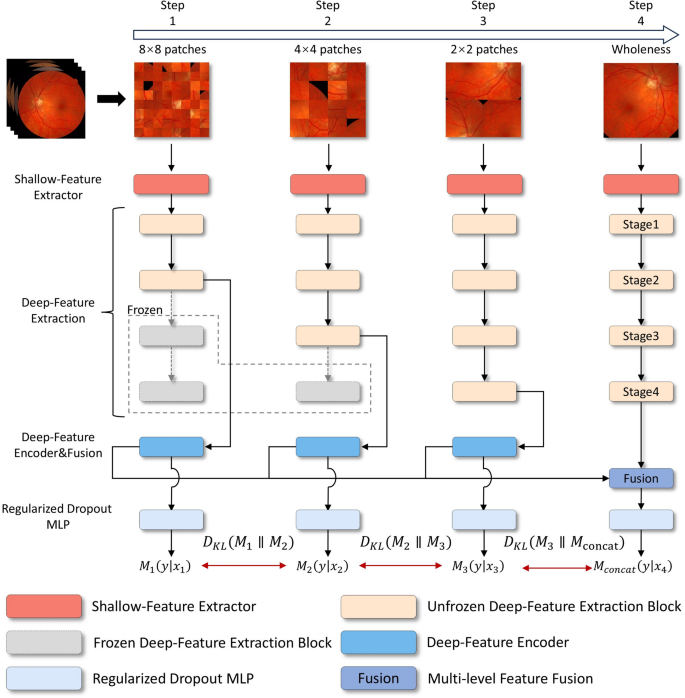
The Diabetic Retinopathy Detection Framework is divided into four steps, each responsible for learning specific levels of detail in retinal images. Guided learning loss is vital in regularization, ensuring that each stage’s classification models do not favor errors.
Each of these feature maps is then fed into its corresponding Regularized Dropout Multilayer Perceptron (MLP), which produces a classification result for each step, yielding a total of four classification outputs. To ensure consistency across these outputs, Kullback-Leibler (KL) divergence is employed as a regularization mechanism (Regularized Dropout Loss (\(\mathcal {L}_{Rdrop}\))) , encouraging the predictions from each classifier to converge towards greater alignment. This design not only enhances the stability of the model but also promotes consistency and robustness in the classification results.
Guided learning loss function
Our GPMKLE-Net framework is optimized using a composite loss function that integrates three distinct components: cross-entropy loss (\(\mathcal L_{CE}\)), class-balanced loss with focal weighting29 (\(\mathcal L_{CB}^{Focal}\)), and regularized dropout30 loss (\(\mathcal {L}_{Rdrop}\)). Each of these loss functions plays a crucial role in improving the model’s performance and robustness.
Cross-Entropy Loss (\(\mathcal L_{CE}\)). This serves as the primary supervised loss term, guiding the classifier to minimize the discrepancy between predicted and actual class probabilities. It provides a solid foundation for the model’s learning by ensuring that the classifier’s predictions align with the ground truth labels.
Class-Balanced Loss with Focal Weighting (\(\mathcal L_{CB}^{Focal}\)). To address the issue of class imbalance in the diabetic retinopathy dataset, a class-balanced loss function with focal weighting has been used. This loss dynamically adjusts weights based on class frequency, assigning greater significance to underrepresented classes29. Additionally, the focal term emphasizes difficult-to-classify samples, further enhancing the model’s performance across all categories. The \(\mathcal L_{CB}^{Focal}\) loss is defined as follows:
$$\begin{aligned} \mathcal L_{CB}^{Focal}= -\frac{1-\beta }{1-\beta ^{n_y}}\sum ^C_{j=1}(1-p_j^t)^\gamma \log (p_j^t). \end{aligned}$$
(1)
In this equation, C represents the total number of classes, and \(n_y\) represents the total number of samples belonging to the target class. The tunable hyperparameter \(\beta\) balances the proportion of positive and negative examples within the dataset. At the same time, \(p_j^t\) refers to the predicted probability of the j-th class, and t represents the actual class for the given sample. Additionally, \(\gamma\) is a tunable factor used to modulate the misclassification loss. By incorporating the class-balancing term \(\frac{1-\beta }{1-\beta ^{n_y}}\) and the focal weighting factor \((1-p_j^t)^\gamma\), \(\mathcal L_{CB}^{Focal}\) effectively addresses two key challenges in medical image classification: class imbalance and varying degrees of classification difficulty.
Regularized Dropout Loss (\(\mathcal {L}_{Rdrop}\)). To enhance both the robustness and generalization ability of the model, regularized dropout techniques are incorporated into the loss function. Specifically, \(\mathcal {L}_{Rdrop}\) imposes a Kullback-Leibler divergence regularization between the output distributions of two distinct classifiers, encouraging their predictions to be consistent30. This guided constraint not only reduces overfitting but also enhances the model’s stability, particularly during the initial stages of training when the model is most susceptible to deviations. Specifically, the \(\mathcal {L}_{Rdrop}\) regularization is applied as follows:
$$\begin{aligned} \mathcal {L}_{Rdrop} = \frac{1}{2} \left( \sum _{j=1}^{C} p_j \log \frac{p_j}{q_j} + \sum _{j=1}^{C} q_j \log \frac{q_j}{p_j} \right) . \end{aligned}$$
(2)
Where C represents the total number of classes, \(p_j\) and \(q_j\) are the predicted probability distributions for the j-th class from the two classifiers, respectively. This formulation ensures that the predictions from both classifiers are in close agreement, effectively reducing the model’s sensitivity to random dropout patterns and enhancing its overall stability. During training, the \(\mathcal {L}_{Rdrop}\) loss is combined with other loss terms within a composite loss function, weighted by a balancing coefficient \(\lambda _{Rdrop}\).
The overall loss function for GPMKLE-Net is a weighted sum of these three loss components:
$$\begin{aligned} \mathcal {L}_{all}=\lambda _{CE} \mathcal {L}_{CE}+ \lambda _{CB} \mathcal {L}_{CB}^{Focal}+\lambda _{Rdrop} \mathcal {L}_{Rdrop} \end{aligned}$$
(3)
By integrating these three loss components through a weighted summation, a comprehensive guided learning loss function is formulated, which serves to systematically direct the model’s learning trajectory. The adjustable hyperparameters (\(\lambda _{CE}\), \(\lambda _{CB}\), and \(\lambda _{Rdrop}\)) allow us to fine-tune the relative importance of each loss term, ensuring that the model is optimized for the specific challenges of DR fundus image classification. A detailed description of the loss function is available in Algorithm 1.

Preprocessing and data augmentation
Given the dataset’s inherent limitations and imbalances, several preprocessing and data augmentation approaches have been implemented.
Dataset extension
Due to the limited availability of manually labeled fundus images, a multi-dataset fusion approach was used, combining various DR fundus image datasets to fully utilize the diversity across data sources and enhance the training set. Enhancements in the generalizability and accuracy of DR detection were pursued by combining these diverse datasets. To guarantee that the combined datasets upheld high quality and consistency, a range of sophisticated data augmentation and image preprocessing methods was methodically applied during the merging process.
Image segmentation
Emphasizing accurate segmentation of fundus images helps minimize the risk of model errors from non-retinal regions. For this purpose, the Otsu thresholding method31, recognized for its efficiency and widespread adoption, was utilized to delineate the retinal foreground from the background. Optimizing the segmentation threshold to maximize inter-class variance led to better detection of retinal boundaries and greatly reduced the effect of dark patches in the images. This step improved the accuracy and quality of fundus region segmentation, contributing to more reliable model predictions.
Image color standardization
Variations in acquisition protocols and environmental conditions frequently lead to color inconsistencies in fundus images, which can significantly impact model performance. To mitigate such effects, image color standardization was undertaken, whereby the mean and standard deviation of pixel intensities across all RGB channels were computed, and each pixel was subsequently normalized in accordance with these statistics. The standardization of color distributions in the training set not only minimized the impact of different data sources but also quickened the model’s convergence.
Histogram equalization sampling
A strategy was created to solve the persistent problem of class imbalance in diabetic retinopathy fundus image classification by integrating histogram equalization sampling with a class-balanced loss function29. Rather than relying solely on conventional sampling methods, histogram equalization sampling was first employed to assess the distribution of samples across categories, with weights subsequently assigned to ensure a more equitable representation among classes. By coupling this sampling technique with a dynamically rescaled class-balanced loss, a synergistic effect is achieved, whereby both model stability and generalization are markedly enhanced. Balanced sampling ensures that underrepresented classes are not neglected during training, and the class-balanced loss further underscores their significance by adaptively changing each class’s contribution to the overall loss. The simultaneous optimization of model parameters and the loss function, directed by these complementary mechanisms, results in a more robust model that is better equipped to deal with imbalanced data distributions.
Dataset
A combination of datasets was employed to ensure both comprehensive analysis and robust model training. The Messidor-1 dataset32, recognized for its high-quality images and precise annotations, served as the primary resource for this study. To address the issue of category imbalance and to enhance data diversity, additional images were selectively incorporated from the Kaggle DR Detection dataset33, with particular attention given to enriching the Mild category.
The Kaggle DR Detection dataset33, originally released in 2015, encompasses a wide range of retinopathy stages—from normal to proliferative retinopathy—captured using various cameras under diverse conditions. While this diversity provides a rich dataset, it also introduces challenges such as noise, overexposure, underexposure, and blurriness due to equipment variations and capture conditions. This dataset’s long-tail distribution of images across categories poses additional complexities in maintaining balanced training sets. Similarly, the APTOS 2019 Blindness Detection dataset34, accessible on Kaggle and divided into five categories, has characteristics akin to the Kaggle DR Detection dataset, such as image noise.
The Messidor-1 dataset32, funded by the French Ministry of Defense’s TECHNO-VISION project, stands out for its precision and quality. Compiled from three ophthalmology facilities, it contains 1200 carefully annotated images. Notably, it is a four-category dataset focusing on diabetic retinopathy. Nevertheless, it was observed that the Mild category is notably underrepresented, a factor that may adversely affect classification accuracy. To mitigate this limitation, 125 high-quality images from the Mild category of the Kaggle DR dataset were strategically selected, thereby enhancing the training set and promoting a more balanced representation of disease stages. The APTOS dataset was further utilized as a validation resource to rigorously assess the effectiveness of the proposed methodological approach. Through this integrated strategy, a comprehensive evaluation of the classification model was achieved, as summarized in Table 1.
Implementation details
In this study, feature extraction was performed using a ResNet-50 backbone35, whose weights had been initialized with ImageNet pre-training36 to mitigate the risk of overfitting, given the limited size of the available dataset. Rather than training a model from scratch, this transfer learning approach was adopted to leverage the generalization capabilities conferred by large-scale pre-training. Training was conducted for 300 epochs. Model optimization was carried out using stochastic gradient descent (SGD) with a batch size of 32, a weight decay of \(5 \times 10^{-4}\), and a fixed momentum of 0.9. The initial learning rate was set to \(8 \times 10^{-3}\) and subsequently annealed according to a cosine schedule, with the minimum learning rate set to \(1 \times 10^{-5}\). Notably, the incorporation of a Guided Loss combination into the loss function represents a methodological innovation unique to this work, whereas all other comparative methods employed the standard cross-entropy loss. To construct the training set, 70% of the dataset was randomly selected, with the remaining 30% reserved for validation and testing. During training, a series of data augmentation techniques were employed to improve generalization, including resizing to 256 pixels, center cropping and random cropping to 224 pixels, random horizontal flipping with a probability of 0.5, random rotations of up to 90 degrees, conversion to tensor format, and normalization using the dataset-specific mean and standard deviation.
All experiments were conducted using the PyTorch framework. Model training and evaluation were performed on an NVIDIA A5000 GPU, ensuring efficient processing of all tasks.