Datasets
Main dataset
A total of 8596 experimentally validated IL-2 inducing and non-inducing peptides were extracted from the largest repository of the immune epitope database (IEDB), filtering for MHC binders from any host organism that was experimentally confirmed to either induce or not induce IL-2 production. Of these, 4475 peptides were MHC binders, which can trigger IL-2 secretion as measured by different immunological assays. These epitopes were termed IL-2 inducing peptides and grouped under a positive set. We also extract 4121 MHC binding peptides that do not trigger the IL-2 secretion and are termed non-inducers. The MHC-binding peptides that do not induce IL-2 are called non-inducers and are grouped under a negative set. Literature evidence suggests that peptides of length between 8 and 25 are most suitable for MHC antigen processing and presentation. Thus, all peptides of length below 8 and above 25 were removed. Additionally, all the redundant peptides were removed. The final main dataset consists of 3429 IL-2 inducing and 3145 non-inducing peptides. One of the major features of our main dataset is all peptides are experimentally validated.
Alternate dataset 1
Our main dataset has all MHC binders, which means models developed on the main dataset are only suitable to MHC binders. In case the user does not know whether a given peptide is an MHC binder or a non-binder, then the model developed on the main dataset cannot be used. In order to overcome this limitation, we changed our negative dataset of non-induces from MHC binders to non-binders. In the Alternate dataset, we extracted and selected 3429 non-MHC-binding peptides from IEDB and assigned them as non-inducers. Finally, our Alternate dataset 1 contains 3429 MHC binding IL-2 inducing peptides as positive peptides and 3429 MHC non-binding IL-2 non-inducing peptides as negative peptides. Models developed on the Alternate dataset 1 are suitable for predicting IL-2 inducing peptides in MHC non-binders.
Alternate dataset 2
Our models developed on our main dataset are suitable for predicting IL-2 inducing peptides in MHC binding peptides. Similarly, our models developed on Alternate dataset 1 are suitable to predict IL-2 inducing peptides in MHC non-binding peptides. In case the user has no idea whether their peptide is an MHC binder or a non-binder, then the above datasets are not suitable. In this study, we proposed alternate dataset 2 that contains 3429 MHC binding IL-2 inducing peptides, referred to as positive peptides and 3429 IL-2 non-induces. In Alternate Dataset 2, IL-2 non-inducers contain a mixture of MHC binders and non-binders, which do not induce IL-2.
Length distribution and composition analysis
To better understand the characteristics of the peptides in both the positive and negative datasets, a comprehensive analysis was performed on their length distribution and amino acid composition. This analysis was carried out using custom Python scripts, which were designed to generate bar plots for visual representation.
Positional preference analysis
In this analysis, a Two Sample Logo (TSL) was constructed to compare the proportional representation or relative abundance of amino acid residues between IL-2-inducing (positive) peptides and non-inducing (negative) peptides. The analysis was focused on identifying the preference for specific amino acids at particular positions within peptide sequences to distinguish between the two peptide groups. Notably, the first eight positions represent the N-terminal residues, while the final eight positions correspond to the C-terminal residues of the peptides.
Alignment-based approach
Motif search
Identification of the motifs within peptides is crucial in annotating their function. The MERCI software was used to identify specific motifs in both positive and negative datasets23. The software was implemented in two steps; firstly, the motifs for a positive dataset were extracted by providing the IL-2 inducing peptides as positive and non-inducing peptides as negative sets. In the second iteration, the motifs were retrieved for the negative dataset by inputting IL-2 non-inducing peptides as positive and IL-2-inducing peptides as negative sets. The motifs were calculated using the earlier approach in both positive and negative datasets. The different classification methods (NONE/BETTS-RUSSELL/KOOLMAN) were used to identify motifs in a mutually exclusive manner with the help of MERCI motif analyses. NONE applies no grouping; BETTS-RUSSELL categorizes amino acids into polar, hydrophobic, and small groups, while KOOLMAN focuses on aliphatic, aromatic, and other properties. The peptides containing unique motifs from both sets were screened to understand the overall coverage of the various motifs in the complete data.
BLAST search
This study utilised blastp-short to annotate IL-2 peptides based on their similarity to IL-2-inducing or non-inducing sequences24. Initially, a database was constructed using the “makeblastdb” command for IL-2 peptide sequences in the training dataset. The self-hits were excluded from the training dataset results and considered the top hit after removing them. The first hit was considered for the independent dataset to calculate results at various e-values.
AI-based classification method
Feature estimation
The different features of peptides present in both positive and negative datasets have been calculated. These calculated features are employed in developing ML-based prediction method development. For the feature generation, the Pfeature web server25 was used, and the embeddings were generated from the pre-trained protBERT model26. With the help of Pfeature, 10,000 peptide descriptors were calculated for both positive and negative datasets. Pfeature includes (I) Composition-based descriptors – Amino Acid Composition (AAC), Dipeptide Composition (DPC), Repetitive Residue Information (RRI), Physico-Chemical Properties (PCP), Distance Distribution of Residues (DDR); (II) Binary profiles – Amino acid based binary profile (AAB); and LLM include Embeddings of Large Language Models trained on ProtBERT developed by Rostlab.
Prediction models
ML models
Various ML-based algorithms have been implemented to classify IL-2 inducing and non-inducing peptides. The “scikit-learn” Python package was used for the classification. The classification algorithm includes – decision trees (DT), random forest (RF), multi-layer perceptron (MLP), eXtreme gradient boosting (XGBoost), support vector with the kernel as a radial basis (SVR), ExtraTreesClassifier (ET) and LASSO. Hyperparameter tuning was implemented using the grid search CV technique with “ROC” as the optimization metric. The DT algorithm was based on the non-parametric supervised algorithm; RF is an ensemble-based method that fits numerous decision trees to predict the outcome of the dependent variable; KNN is an instance-based learning algorithm, and XGBoost is a tree-boosting classification algorithm based on an iterative search approach for making the final prediction.
DL models
For the DL approach, two distinct models were applied: first, a 1D CNN model tailored for processing sequential data and extracting hierarchical features along the sequence, and second, TabNet, a modern neural network architecture designed explicitly for tabular data27. The 1D CNN Model is particularly effective for sequential data processing, capturing patterns and dependencies along the peptide sequences. On the other hand, TabNet excels in tabular data by dynamically selecting and reasoning from features using sequential attention, making it suitable for our IL-2 inducing and non-inducing peptides dataset. Each model was fine-tuned independently on the datasets by adjusting hyperparameters to optimize their performance for the classification task.
LLM models
LLM are excellent at text classification because they deeply understand language. Trained on large amounts of text, they can be designed for specific tasks like classifying sequences, as was done in this research. LLMs are flexible and are particularly good at extracting important text features, allowing them to make accurate classifications even with limited data. This study used LLMs to differentiate between IL-2 and non-IL-2 sequences. The protBERT model was chosen and fine-tuned to match the characteristics of the datasets. After this fine-tuning, the model predicted whether each sequence belonged to the IL-2 or non-IL-2 category.
Feature selection
As the literature suggests that not all descriptors are effective for developing machine learning models, three techniques were employed to select the best features: minimum Redundancy – Maximum Relevance (mRMR), SVC-L128 and Recursive feature elimination (RFE). Feature selection was performed on DPC with length. The top 10, 100, and 200 features were computed using both methods on the datasets. After the feature selection step, all the features mentioned above were tested independently.
Ensemble method
In this approach, the best model obtained was combined with motif information from MERCI for reliable and biologically significant prediction of IL2-inducers. Along with the prediction models, the query peptide was searched against the test data with different thresholds for false positive (fp) values; if any motif was found in the peptide, the probability score of test data was increased by 0.5 for IL-2 inducing peptides and in the case of IL-2 non-inducers, the score was decreased by 0.5. This gives the probability of a sequence belonging to a specific class instead of a binary outcome.
Cross-validation
For developing the ML, DL, and LLM models, the standard protocols were followed as in previous studies [31498794]. Each dataset was divided into an 80:20 split for training and external validation, respectively. The main dataset contained 5,259 peptides for training and 1,315 peptides for testing. The alternate dataset 1 included 5,486 peptides for training and 1,372 for testing, while the alternate dataset 2 consisted of 5,487 peptides for training and 1,372 for testing. The different classifiers were trained and evaluated using a five-fold cross-validation method, a widely accepted technique for optimizing model parameters and performance. In the five-fold cross-validation, the training dataset was divided into five equal parts; iteratively, four parts were used for training and one for testing to fine-tune the model parameters. This process was repeated five times, ensuring each subset was used for both training and testing. All the classifiers were implemented using an in-house python script. Performance evaluation was based on metrics such as specificity, sensitivity, accuracy, MCC, and AUC. The statistical performance-evaluation parameters of the ML models are explained below:
$$\text{Sensitivity} =\:\frac{TP}{\left(TP+FN\right)}$$
(1)
$$\text{Specificity} =\:\frac{TN}{\left(TN+FP\right)}$$
(2)
$$\text{Accuracy} = \:\frac{TP+TN}{\left(TP+TN+FP+FN\right)}$$
(3)
$$\text{MCC}=\:\frac{TP\times\:TN-FP\times\:FN}{\sqrt{\left(TP+FP\right)\left(TP+FN\right)\left(TN+FP\right)\left(TN+FN\right)}}$$
(4)
Architecture of Web-server
A web server named “IL2pred” (https://webs.iiitd.edu.in/raghava/il2pred/) was developed to predict the IL-2 inducing and non-inducing peptides. The web server’s front end was developed using HTML5, JAVA, CSS3, and PHP scripts. It is based on responsive templates that adjust the screen based on the device’s size. It is compatible with almost all modern mobile, tablet, and desktop devices. The overall workflow of the complete methodology is summarized below in Fig. 5.
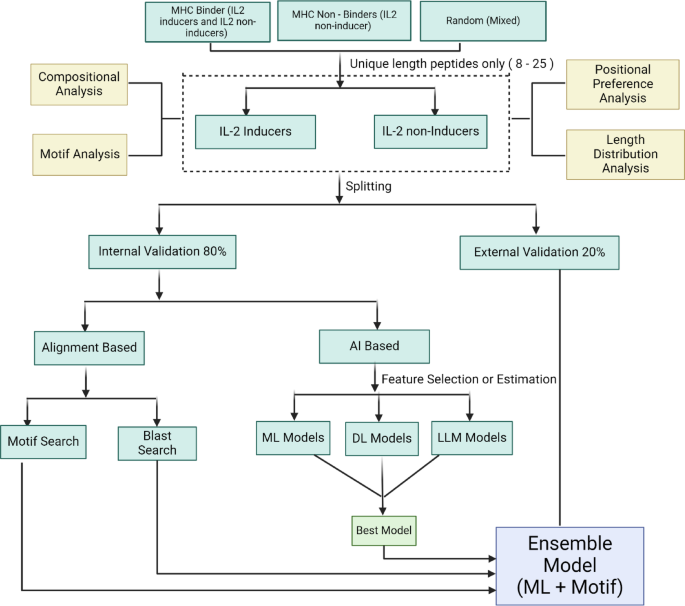
Schematic representation of the workflow of the study.
Modules and functionalities of IL2pred web-server
This web server is compatible with all sorts of devices, viz. desktops, tablets, and phones, and hence provides an interactive and better experience to the users. Built with Python 3.12.2, the server utilizes essential scientific libraries including scikit-learn (v1.5.2), Pandas, and NumPy for efficient computational operations. The webserver has four main modules – (1) Prediction; (2) Design; (3) Protein Scan; (4) Motif Scan. The “Prediction module” allows users to identify the peptides with IL-2 inducing potential. Users of the web server can submit the peptides of interest in FASTA format as well as upload the FASTA format files from their local machine. The results of the user-defined query are displayed on the HTML page as IL2-inducer and IL-2-non-inducer, along with the prediction score and selected properties. The results of the user-defined query can also be downloaded from the web server in the CSV format. The “Design module” is of utmost importance for a biologist who wishes to design new, potent peptides with enhanced IL-2 inducing potential. The input for the design module is a single-line peptide sequence file, which can either be pasted in the box provided on the module page or locally uploaded onto the web server site. Once the user-defined query is submitted on the web server, the algorithm implemented on the “Design Module” generates all possible mutants of the given query peptide sequence and then ranks them based on their IL-2 inducing potential. The ranking algorithm is of greater importance as it ranks the newly generated single-letter mutant peptides for their ability to induce IL-2. Thus, it helps biologists narrow down their choice of selecting the best mutant peptide with enhanced IL-2 inducing ability. The prediction score and the other predicted metrics are displayed on the web server site, which can be downloaded as a CSV file for further use. The “Protein Scan” module lets users predict IL-2 inducing regions within a given protein sequence. Users can submit the protein sequence of their interest in a single-line format. The algorithm generates all possible fragments of the user-defined length and then predicts the IL-2 inducing potential of all generated fragments overlappingly. This will help in the identification of IL-2 rich and scarce regions in a given protein sequence and thus help biologists in selecting and prioritizing appropriate regions of protein for their study the last model is the Motif scan, which will allow users to find crucial motifs present in the query protein sequence to find out its role as an IL-2 inducer or IL-2 non-inducer. The results from this module can be downloaded in CSV format. The user-friendly web server, along with pip package and standalone maintained in GitHub, is available at (https://webs.iiitd.edu.in/raghava/il2pred ).