Researchers at Seoul National University have developed a powerful new computational tool called T-DNAreader that significantly advances the study of genetically modified plants. In plant research, one of the most common methods to introduce new genes into plants is through Agrobacterium-mediated transformation. This process uses a natural soil bacterium, Agrobacterium tumefaciens, which transfers a piece of DNA known as transfer DNA or T-DNA into the plant genome. Scientists exploit this system to insert desired genes, which can help improve crop traits such as disease resistance, drought tolerance, or yield.
However, a major challenge in working with genetically modified plants is identifying the exact locations where these T-DNAs insert into the plant’s DNA. The T-DNA integrates randomly in the genome, and sometimes multiple T-DNA sequences can insert at once during a single transformation event. Knowing precisely where these insertions occur, called T-DNA insertion sites or TISs, is critical for understanding how the inserted genes behave, whether they disrupt important native genes, and for verifying the genetic modification itself.
To address this challenge, the team developed T-DNAreader, a tool that uses RNA sequencing data to locate T-DNA insertion sites with high accuracy, speed, and sensitivity. RNA sequencing is a technique that reads the actively expressed genes in a cell by sequencing their RNA transcripts. By analyzing this data, T-DNAreader can identify where T-DNA sequences appear in the transcripts, effectively pinpointing where the DNA has inserted within the parts of the genome that are actively being used by the plant.
Workflow of T-DNAreader
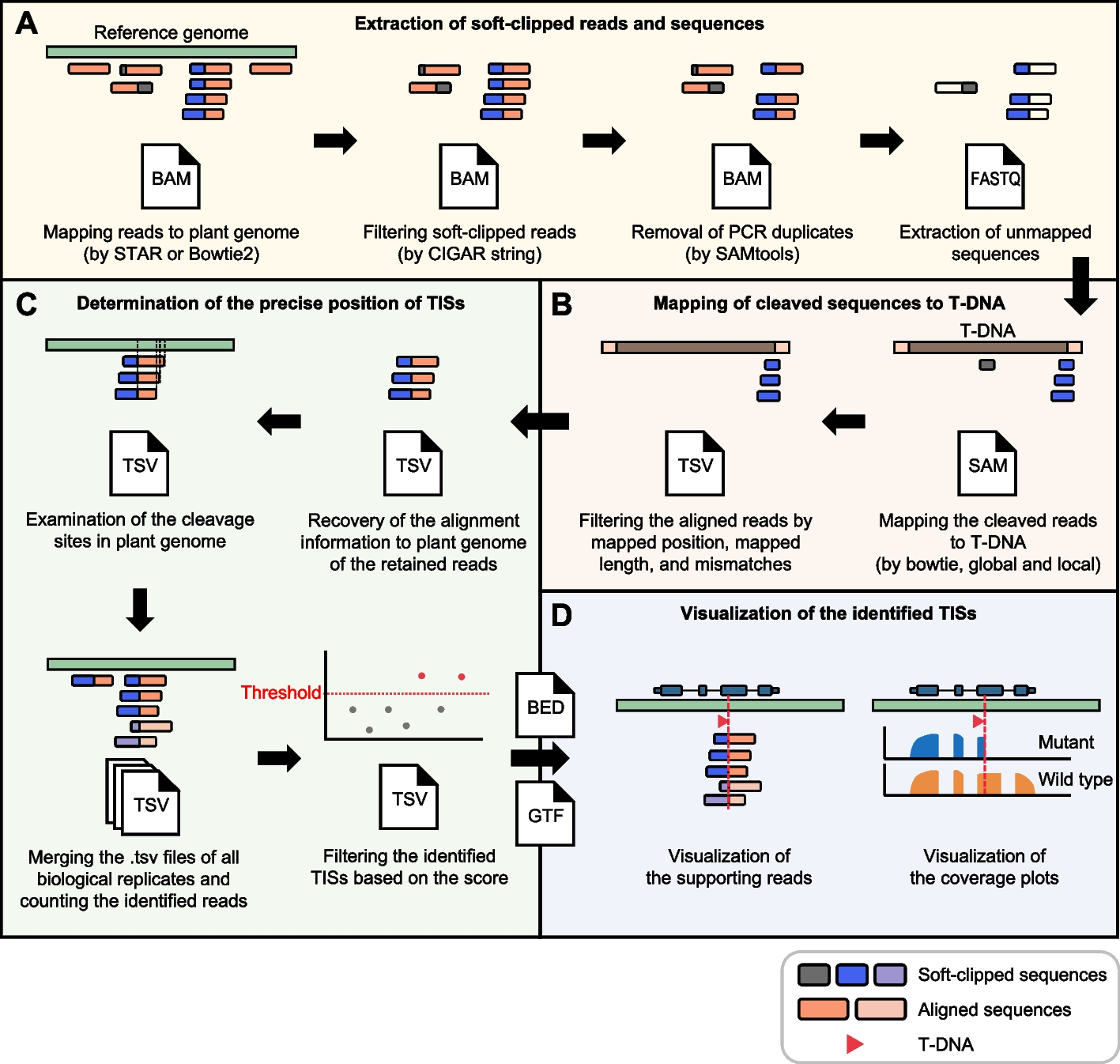
The T-DNAreader pipeline consists of four processes. A FASTQ files are aligned to the reference genome, and then soft-clipped reads are collected from aligned BAM files based on the CIGAR string “S”. After removing duplicate reads, new FASTQ files containing the unmapped portions of the soft-clipped reads are then generated (cleaved FASTQ files). B The cleaved FASTQ files are mapped to the provided T-DNA sequences. Reads successfully mapped to the T-DNA sequences are filtered based on (1) their alignment position within the T-DNA, (2) the length of mapped sequences, and (3) the number of mismatches allowed. C The original alignment of the retained reads to the plant genome is restored, and their exact cleavage sites are identified. The information from all biological replicates is merged, and a confidence score is calculated for each candidate TIS. The TISs with scores above a defined threshold are retained. D T-DNAreader generates visualizations for each identified TIS, including supporting reads and coverage plots. The genomic positions and orientations of the T-DNA insertions are shown with the corresponding genomic annotations. In A–D, T-DNA and genomic segments of chimeric reads, and soft-clipped reads originating from different replicates are displayed in distinct colors
What makes T-DNAreader especially valuable is its ability to process large datasets quickly while maintaining precision. It outperforms existing tools that often require DNA sequencing or more complex procedures. Since it focuses on transcribed regions, T-DNAreader is particularly useful for researchers who want to understand the functional impact of the T-DNA insertions on gene expression.
This method not only speeds up the process of characterizing genetically modified plants but also provides a standardized way to analyze T-DNA insertions. Such standardization is essential for comparing results across different experiments and laboratories. Moreover, while the tool was developed for plants, its application can be extended to other organisms that have been genetically modified using T-DNA.
Overall, T-DNAreader represents a significant step forward in plant genomics and biotechnology, offering researchers a reliable and efficient way to map T-DNA insertions. This will help accelerate research in crop improvement, gene function analysis, and the development of new biotechnological tools for agriculture.
Availability – The source code of T-DNAreader is freely available on GitHub (https://github.com/CDL-HongwooLee/TDNAreader)