Transposable elements, or TEs, are fragments of DNA that can move around the genome. Once thought to be genomic “junk,” scientists now understand that these mobile genetic elements influence everything from embryonic development to neurological disorders and cancer. Surprisingly, TEs account for nearly half of the mammalian genome, yet studying their function has remained a significant challenge, especially at the single-cell level.
One reason for this difficulty is that many TEs have nearly identical sequences. When researchers use short-read RNA sequencing, a common method that captures only small snippets of RNA, they often can’t tell exactly where a TE read came from. That ambiguity makes it hard to track TE activity precisely in individual cells.
Researchers at the University of Oxford introduced an innovative solution: CELLO-seq (Cellular Long-read sequencing). This protocol combines the power of long-read sequencing with a clever barcoding technique to make it possible to map TE-derived RNA reads back to their exact location in the genome, even when sequences are highly similar.
Schematic representation of the CELLO-seq wet lab workflow labeled with key steps in the Protocol
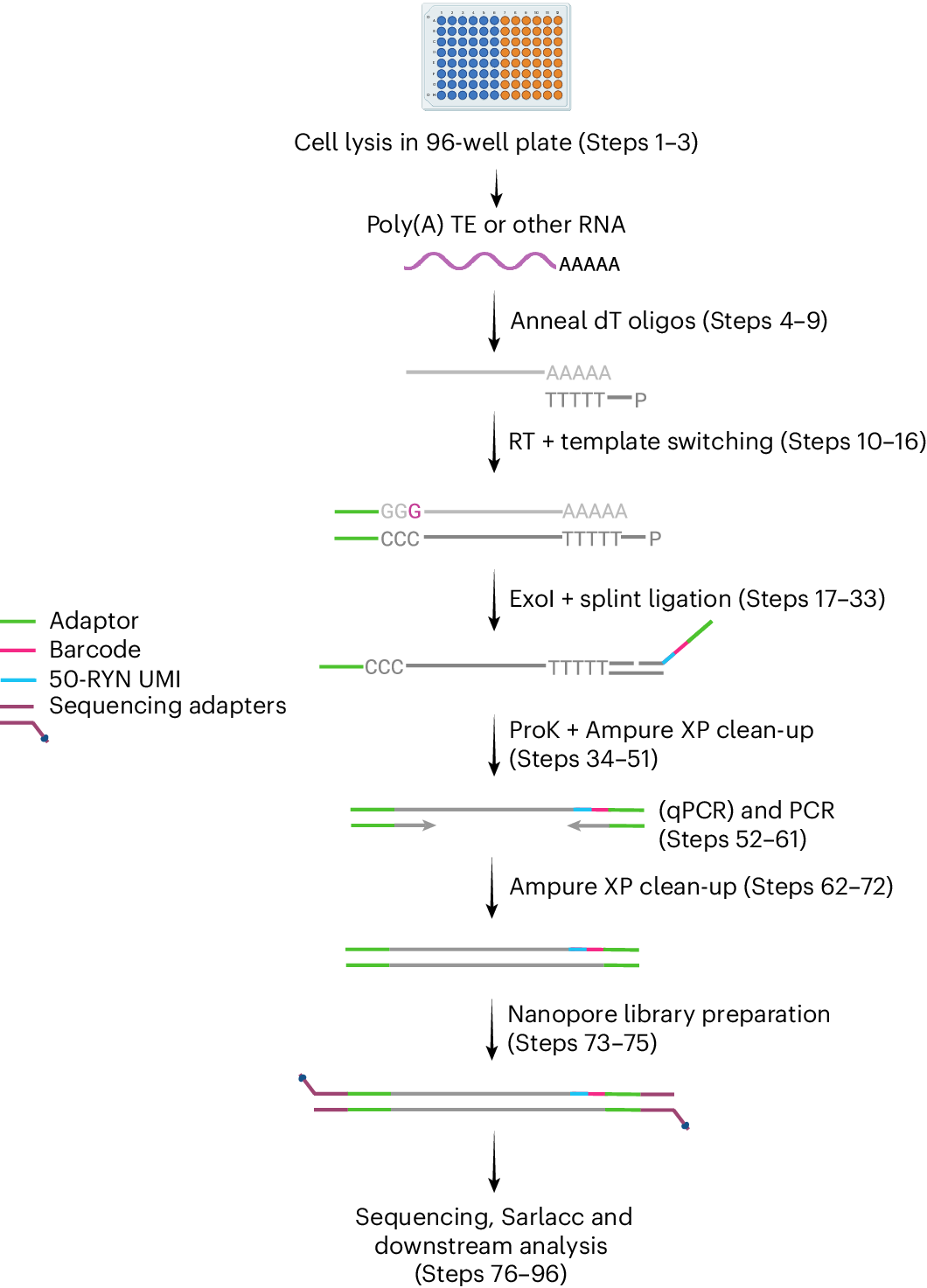
The single cells for CELLO-seq are first sorted into individual wells of a 96-well plate containing lysis buffer. The dT oligos are annealed to the poly(A) tail of RNAs, priming the RT and template switching. Following the ExoI treatment, the splint oligos are ligated, consisting of an adaptor, barcode and 50-RYN UMI (green, pink and blue, respectively). The proteinase K digestion of contaminant proteins is followed by an AMPure XP cleanup and PCR. After the additional AMPure XP clean-up steps, the ONT library preparation can be performed, whereby ONT sequencing adaptors (purple) are ligated onto the amplified cDNAs. The ONT library is sequenced on a MinION or PromethION flow cell, and the fastq files are analyzed with the Sarlacc pipeline.
CELLO-seq works by tagging each RNA molecule with a long unique molecular identifier (UMI), a 50-nucleotide barcode, before amplification. This enables scientists to group and correct sequencing errors by comparing multiple reads from the same original RNA molecule. The method also involves generating high numbers of PCR duplicates, which further aids in error correction and data accuracy.
The result? CELLO-seq can confidently identify not only where each transposable element is expressed in a single cell but also which isoforms of TEs and genes are being produced. This is especially useful for studying young, active TEs that are more likely to impact gene regulation or disrupt cellular function.
This protocol represents a significant leap forward in our ability to study the complex landscape of gene regulation. By making it possible to look at TE activity in individual cells, CELLO-seq offers new insights into how these elements contribute to biological diversity, development, and disease.
Designed for researchers already familiar with RNA workflows and transcriptomic analysis, the CELLO-seq method can be completed in about a week, from isolating cells to analyzing the sequencing data. As more labs adopt this technique, we can expect to learn much more about the “dark matter” of the genome and how it influences life at the cellular level.